In Search of New “Sugar Cleavers”
Complex sugars play multiple and essential roles in the living world as structural elements (cellulose), reserve substances (starch), and molecular signals (such as those of blood types), among others. This variety of functions is based on structures that are equally diverse, due to a multiplicity of building blocks and the different ways in which they can be arranged. The degradation of these carbohydrates therefore requires a variety of specific enzymes, which are far from being identified in their entirety, especially in the microbial world: the human genome contains only a dozen, but our intestinal microbiota has 60,000–70,000! Two French teams from the CNRS have developed a method for accelerating the discovery of these enzymes, and have subsequently identified 79 new ones, in addition to 13 new families (whereas less than 200 families had been described in over a century). Using bioinformatics methods, scientists from the laboratoire Architecture et fonction des macromolécules biologiques (CNRS/Aix-Marseille Université) looked within the thousands of genomes1 for genes that can code these enzymes. They then produced the protein corresponding to 560 of these sequences. Researchers from the Centre de recherches sur les macromolécules végétales, a CNRS lab in Grenoble, took over from there in an effort to identify the function of these enzymes, by exposing them to a collection of over 200 complex sugars. Despite the newly discovered ones published this week in PNAS, there are still 243 proteins whose activity has not yet been established. Aside from the better understanding of life that they enable, these enzymes can serve as tools in various domains, from bioenergy to cosmetics and nutrition.
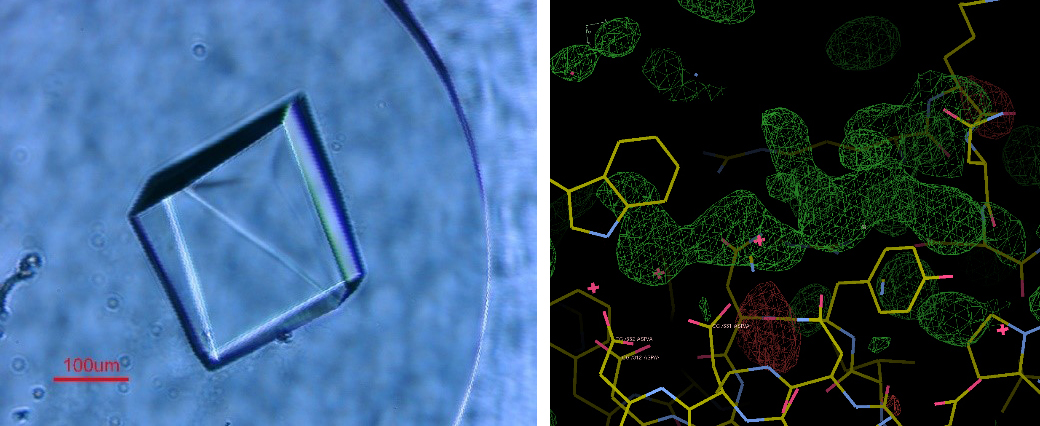
© Marie-Line Garron/AFMB/CNRS-AMU
To find out more: Sugars, New Molecules for Health, CNRS News, 28 December 2018.
- 1Primarily bacterial.
Discovery of novel carbohydrate-active enzymes through the rational exploration of the protein sequences space, William Helbert, Laurent Poulet, Sophie Drouillard, Sophie Mathieu, Mélanie Loiodice, Marie Couturier, Vincent Lombard, Nicolas Terrapon, Jeremy Turchetto, Renaud Vincentelli et Bernard Henrissat. PNAS, 4 March 2019. DOI: 10.1073/pnas.1815791116


